ABOVE: NIAID
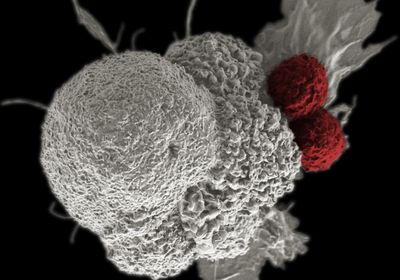
New research suggests that transposable elements—or so-called “jumping genes”—may help scientists identify tumor-targeting proteins. The study, published Monday (March 27) in Nature Genetics, suggests that during cancer, transposable elements make their way into genes and can then be translated into tumor-specific antigens, which are proteins on the tumor surface that mark the cell for immune destruction. The findings may help researchers design cancer therapies that are more generalized across patients that have a particular cancer type than current treatments, the study authors suggest.
Transposable elements (TEs) are short, repetitive sequences that can copy themselves into other regions of the genome. Normally, TEs are epigenetically silenced. But when a cell becomes cancerous, they can begin to hop to different regions of the genome. According to the study, many tumor-specific antigens stem from when a transposable element inserts into the coding region of a gene, making its way to the final protein product.
“It has been shown before that some transposable elements are translated and can be presented to cells of the immune system. What was impressive was the scope of the analysis,” says Claude Perreault, an immunogeneticist at the University of Montreal, who was not involved with the study. “I think they did superb work.”
Study co-author Ting Wang, a geneticist at Washington University School of Medicine in St. Louis, didn’t start off studying cancer. Instead, he had long been interested in how transposable elements contribute to evolution, particularly in the context of regulatory elements. Then, five years ago, he and his lab made several discoveries about how transposable elements disrupt regulatory elements during cancer, which he calls “a different form of evolution.”
See “Adapting with a Little Help from Jumping Genes”
From the lab’s previous work, he knew that when transposable elements destabilize regulatory elements, they allow the expression of regions of the genome that would normally lie dormant. Occasionally, this aberrant transcriptional regulation results in a chimeric transcript, wherein a transposable element tacks itself onto a normal gene, resulting in a mashup of a regular gene and a jumping gene, Wang explains.
Although the exact mechanism behind this phenomenon is unclear, transposable elements often lie in introns, which may fail to be spliced out. Researchers had observed this phenomenon previously, but they’d yet to characterize how common it was—which is what Wang set out to do.
For the study, Wang and colleagues analyzed data from The Cancer Genome Atlas (TCGA), a large, open-source database containing genetic sequences of various human tumor samples. Using bioinformatics approaches, the scientists searched mRNA sequencing data from more than 10,000 samples in 33 tumor types—as well as corresponding control tissues—for chimeric transcripts.
The approach was the reverse of a typical mRNA sequencing analysis, Wang explains. “Typically, people take RNAseq data to map genes and understand the gene expression level,” he says. “But in this case, what we’re hoping to do is to discover a novel gene product. So instead of mapping the data to the genes, we made the transcript,” then mapped the novel transcript to the data.
The results shocked Wang. Across all cancer types, he and his colleagues found roughly 2,300 different chimeric transcripts that were unique to tumors—many more than they expected. In addition, 98 percent of tumors had at least one chimeric transcript event. Many of these chimeric transcripts were shared across tumors of a single cancer type.
But that only told the researchers that the chimeric transcript was being transcribed, not translated. So, using publicly available cancer cell line data from the Genentech cell line bank, another publicly available dataset, the team searched the available mRNA sequencing data to find chimeric transcripts in cancer cell lines. Once they found these transcripts, they predicted that some would be regulatory elements, as they’d found in their previous studies. Using nanoCAGE sequencing, a method of genome-wide promoter analysis, they indeed found that some of the chimeric transcripts provide tumor-specific promoters.
Then, they went on the hunt for tumor-specific antigens. After obtaining the cell lines in the lab, they used MHC pull-down, which is a technique for isolating proteins on the tumor cell surface. Then, they identified the antigens with mass spectrometry, finding evidence that the tumor-specific antigens were translated from chimeric transcripts.
See “Jumping Genes’ Role in Cancer”
Wang mentions that the authors have not yet tested whether the proteins directly cause an immune response. However, Perreault says he would be “optimistic” because other groups have found transposable element-derived proteins to be immunogenic. He also hopes that the authors will expand their analyses to other cell lines and primary tumors in the future.
Wang hopes that the findings will improve cancer therapies down the road, as immunotherapies such as CAR-T cell therapy and cancer vaccines rely on tumor-specific antigens, but says that there are many necessary steps before this becomes a reality. Identifying specific mutations is effective in cancers with a high mutation load, such as melanoma, but less effective in other cancers with a low mutation rate, such as glioblastoma and pediatric cancers. Chimeric transcripts are an abundant source of new antigens, sometimes more abundant than mutation-driven tumor-specific antigens, he says.
“Mutation-driven events are often very patient-specific,” says Wang. According to the research, chimeric transcription events are much more recurrent across tumors of the same cancer type, which could perhaps allow researchers to design therapies that work for multiple patients with the same cancer.
“A new antigen is a big deal,” says Wang. “Immunotherapy has truly changed how we treat cancers. A lot of immunotherapies rely on the identification of new antigens, which is why people sequence the cancer genome to try to identify mutations that give new antigens.”