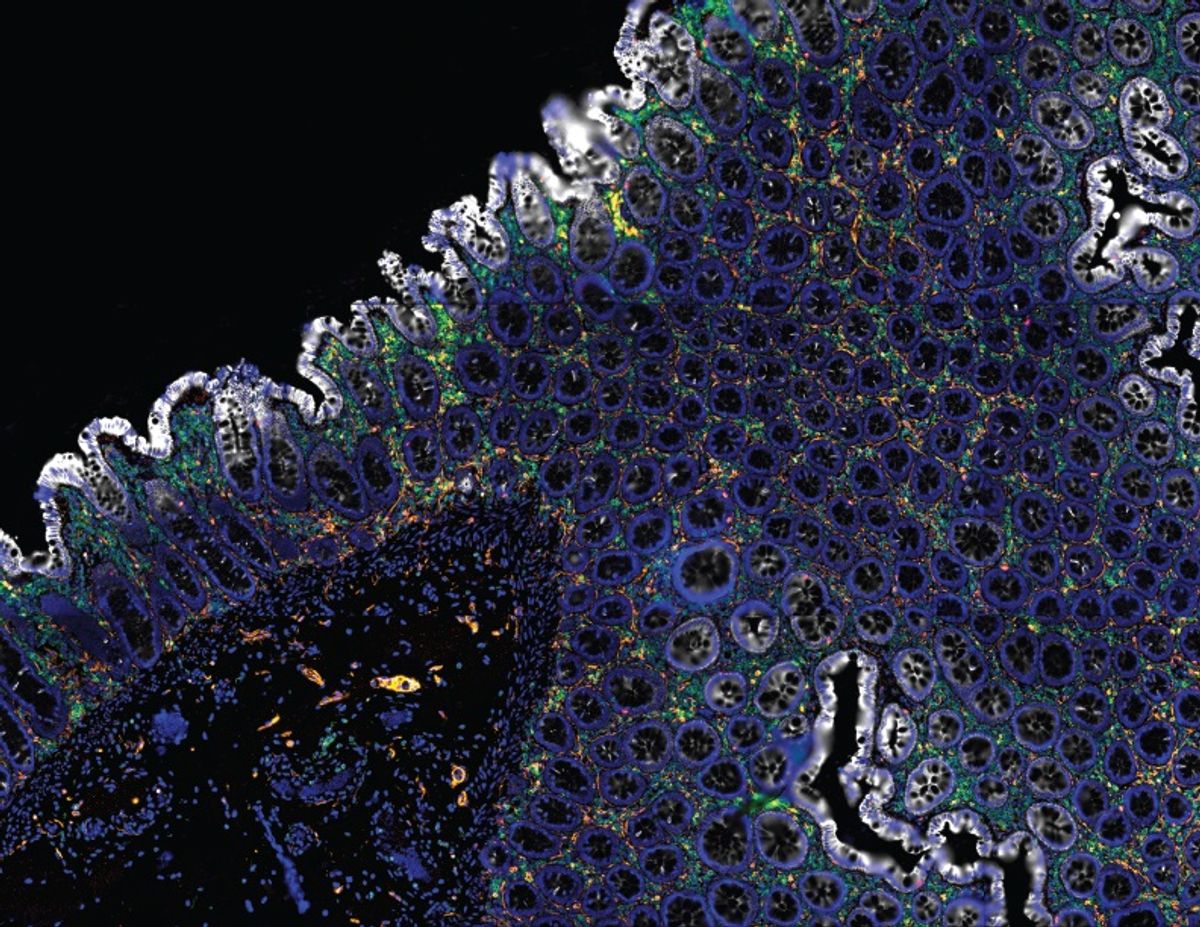
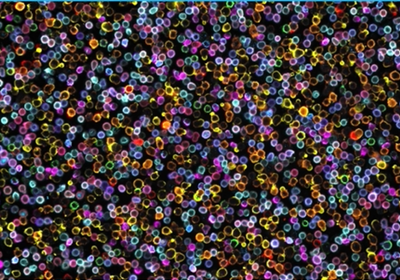
The last decade has heralded the spatial single cell -omics era. Using new techniques, researchers build comprehensive cellular and molecular maps by detecting numerous transcripts and proteins at single cell resolution and in three-dimensional space. Many spatial single cell -omic techniques focus on mRNA profiling within a tissue space.1 While transcriptomic analysis distinguishes cell subtypes and identifies cell states, researchers must corroborate these results with protein analyses to recognize unique molecular signatures present in individual cells. Exploiting such spatial proteomic datasets allows scientists to classify cells, identify novel cell types and genes, trace cell lineages, and uncover causative disease pathways.
Researchers typically want to profile as many protein markers in a sample at once to obtain a better picture of tissue organization and function. One way to visualize protein expression in subcellular compartments is with fluorescent immunohistochemistry (IHC), a powerful approach for characterizing biologically-relevant cellular phenotypes. Traditional fluorescent IHC is an inherently low-throughput and laborious method; staining a few protein markers within tissue sections can take up to a week.2 Profiling only a limited number of markers in each sample at a time limits scientists’ ability to capture the complexity in a tissue, and therefore, reduces the scope of discovery. Moreover, the subsequent image acquisition and data analysis are long processes that require a skilled person to operate a sophisticated microscope. Therefore, researchers developed multiplexed tissue imaging approaches that simultaneously process and analyze numerous markers in thousands of cells.
Immunohistochemistry Meets Automation and Multiplexing
Recent advancements in electronic, optical, and chemical technologies made multiplexed cellular imaging a reality by enhancing experimental speed, scale, resolution, and throughput. CellScapeTM ChipCytometryTM is a revolutionary platform that combines an advanced optical imaging system with fluidics for multiplexed spatial applications, allowing scientists to distinguish unlimited protein biomarkers in a tissue sample and quantify expression of those markers with single-cell resolution. Equipped with an automated liquid handler, slide scanners, and an imaging pipeline for around the clock image acquisition, it processes multiple samples without constant user intervention.
The CellScape automated workflow involves three steps to achieve multiplex tissue staining. First, the immunostaining step labels tissue samples with up to 5 fluorescently-labeled antibodies at a time. Next, the imaging step uses high dynamic range (HDR) imaging to acquire images for a broad range of low- and high-expression targets with superior accuracy and resolution. Lastly, the photobleaching step removes fluorescence signals, which enables immunostaining of the same sample with a new set of antibodies.
The HDR image acquisition feature makes CellScape a unique imaging platform that accurately quantifies low- and high-intensity signals, allowing researchers to profile low- and high-expressing target proteins simultaneously. By capturing images with different exposure times, the system detects a broad range of signal intensities and combines them to create a single composite image. Additionally, the HDR system automatically removes autofluorescence and retains image acquisition metadata for the user to perform advanced analyses. Lastly, the CellScape data analysis tool features automatic cell segmentation and statistical analysis for generating dot plots for expression values easily and efficiently.
Compared to other multiplex tissue imaging platforms, CellScape works with widely-used published and open-source antibody reagents, making the application cost-sensitive as well as easy to adapt and validate in individual laboratories. Using the system, researchers can standardize a wide range of antibodies from different vendors, which makes the instrument applicable to a range of research fields, including neuroscience, oncology, immunology, and infectious diseases.
Spatial Phenotyping in Human FFPE Tissue
Biologists use animal models to understand anatomy, physiology, and disease mechanisms; however, there is no substitute for the unparalleled insight gained from human tissue. Most clinically-relevant human tissue samples are preserved as formalin-fixed paraffin-embedded (FFPE) sections. Recently, researchers used ChipCytometry on healthy and cancer FFPE tissue slices to investigate immune cell infiltration in tumors.3 They successfully located 30 immune cell markers in a variety of colon, breast, and pancreas tissue sections, quantified the number of immune cells, and identified rare cell types. With a continuous increase in the number of commercially available, fluorescently-conjugated antibodies for FFPE tissue sections, ChipCytometry has the potential to revolutionize clinical diagnostics research.
References
- L. Waylen et al., “From whole-mount to single-cell spatial assessment of gene expression in 3D,” Commun Biol, 3:602, 2020.
- L.L. Matos et al., “Immunohistochemistry as an important tool in biomarkers detection and clinical practice,” Biomark Insights, 5:9-20, 2010.
- S. Jarosch et al., “Multiplexed imaging and automated signal quantification in formalin-fixed paraffin-embedded tissues by ChipCytometry,” Cell Rep Methods, 1(7):100104, 2021.