What is DNA Methylation?
DNA methylation is a fundamental epigenetic mechanism in which a methyl group is added onto a nucleotide, commonly cytosine. DNA methylation affects many biological processes, including gene expression, embryonic development, inflammation, and cellular proliferation and differentiation. Several complex diseases have aberrant DNA methylation patterns, such as different types of cancer and neurodegenerative disorders, which are often associated with genomic instability and loss of DNA homeostasis.1–3
Researchers need effective methods with high sensitivity and reliability to explore the importance of DNA methylation. The gold standard technology that scientists use to detect DNA methylation is bisulfite genomic sequencing. It is a qualitative, quantitative, and efficient approach to identify methylated cytosine at single base-pair resolution.3
Bisulfite Conversion of Methylated Cytosine
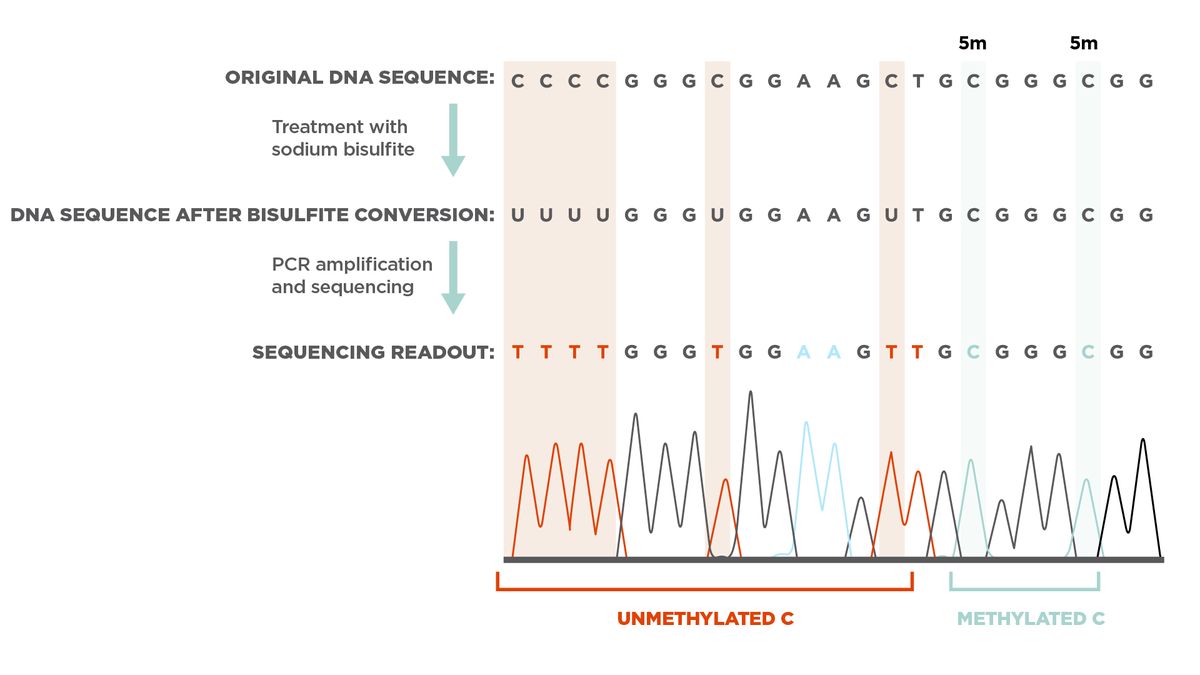
Cytosine methylation occurs when a methyl group binds to the fifth carbon of a cytosine to form 5-methylcytosine (5mC). Researchers cannot detect 5mC with traditional molecular techniques such as PCR or cloning methods because methyl groups are not copied during PCR amplification. Instead, researchers must treat the methylated DNA samples with bisulfite prior to detection, to distinguish between methylated and unmethylated cytosines.1,2
Bisulfite conversion is the chemical reaction that occurs when scientists treat DNA with sodium bisulfite. In 1992, researchers found that the amination reaction of sodium bisulfite with unmethylated cytosine is different than the reaction of sodium bisulfite with 5mC. Because of this difference, unmethylated cytosines in single-stranded DNA become uracil residues after exposure to sodium bisulfite, while 5mCs remain cytosines. Pretreatment with sodium bisulfite is the basis of many methylation detection and analysis techniques.1 Following bisulfite conversion, researchers determine the methylation status in loci of interest because unmethylated cytosines are recognized as thymine after PCR amplification and sequencing.3
Challenges of Treating DNA with Bisulfite
Incomplete bisulfite treatment is a major challenge in methylation detection methods. Bisulfite conversion relies on the chemical modification of cytosine in single-stranded DNA, so scientists must ensure that their DNA sample is completely denatured prior to sodium bisulfite treatment. As such, the quality and quantity of purified DNA and the pH of the reaction are fundamental sample parameters in complete bisulfite conversion.1 Incomplete conversion typically leads researchers to overestimate the amount of methylation in a sample.2 An additional challenge to bisulfite-based methylation analyses is the sensitivity of the DNA sample to degradation during long incubation steps.1
Types of Bisulfite Sequencing
Whole Genome Bisulfite Next-Generation Sequencing
Methylation detection methods are diverse in accuracy, sensitivity, speed, simplicity, and cost. Researchers investigate the methylation of an entire genome, also referred to as a methylome, with whole genome bisulfite sequencing (WGBS). WGBS consists of library preparation with bisulfite treatment, next-generation sequencing (NGS), and high throughput analysis in which researchers explore the biological processes connected to the observed methylome.1,2,5 Challenges during WGBS arise from protocol changes that cause discrepancies in coverage depth, read quality, duplication rates, mapping efficiency, and methylation estimation. Additionally, the standard WGBS protocol requires a large amount of DNA, which may be a roadblock in many methylome studies.2,5
Reduced Representation Bisulfite Sequencing
An alternative approach to WGBS is reduced representation bisulfite sequencing (RRBS). This method reduces the cost and complexity of methylome analysis. With RRBS, scientists enrich for GC-rich parts of the genome by digesting the DNA with an enzyme that generates fragments with CpG dinucleotides at both ends, independent of methylation status. This enrichment is informative because DNA methylation exists primarily in the context of symmetric CpG dinucleotides. After digestion, researchers use size selection to isolate short DNA fragments that correspond to GC-rich regions. Larger fragments that are not GC-rich and contain fewer potential methylation sites are discarded. This digestion and selection step essentially excludes uninformative DNA prior to bisulfite treatment, while capturing the majority of relevant genomic regions of the methylome. RRBS reduces the number of reads needed to obtain high coverage and requires much less DNA than WGBS, but researchers can only examine a fraction of the methylome with this technique due to the amount of excluded DNA.4–7
Targeted Bisulfite Sequencing
In contrast to the broad enrichment of GC-rich regions with RRBS, targeted bisulfite sequencing allows researchers to investigate the methylation status of each cytosine in a specific genomic region. Scientists apply targeted methods to validate differentially-methylated regions or analyze candidate regions rather than the entire methylome. Targeted bisulfite sequencing relies on two PCR amplification steps after bisulfite conversion, followed by deep sequencing. The PCR steps increase copy number and introduce adaptors required for sequencing as well as sample-specific identifiers. These adaptors and identifiers allow researchers to process a large number of samples simultaneously on a high-throughput instrument.7,8
References
- R. Halabian et al., “Laboratory methods to decipher epigenetic signatures: a comparative review,” Cell Mol Biol Lett, 26:1-30, 2021.
- T. Gong et al., “Analysis and performance assessment of the whole genome bisulfite sequencing data workflow: currently available tools and a practical guide to advance DNA methylation studies,” Small Methods, 6:e2101251, 2022.
- Y. Li, T.O. Tollefsbol, “DNA methylation detection: bisulfite genomic sequencing analysis,” Methods Mol Biol, 791:11-21, 2011.
- Y. Zhang, A. Jeltsch, “The application of next generation sequencing in DNA methylation analysis,” Genes (Basel), 1:85-101, 2010.
- Q. Wang et al., “Tagmentation-based whole-genome bisulfite sequencing,” Nat Protoc, 8:2022-32, 2013.
- H. Gu et al., “Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling,” Nat Protoc, 6:468-81, 2011.
- D.A. Moser et al., “Targeted bisulfite sequencing: A novel tool for the assessment of DNA methylation with high sensitivity and increased coverage,” Psychoneuroendocrinology, 120:1-8, 2020.
- E. Leitão et al., “Locus-specific DNA methylation analysis by targeted deep bisulfite sequencing,” Methods Mol Biol, 1767:351-66, 2018.