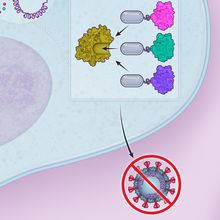
ABOVE: © ISTOCK.COM, DESIGN CELLS
During an early COVID-19 lockdown in March 2020 in Copenhagen, Denmark, we, like many in the scientific community, turned our attention to SARS-CoV-2, the virus responsible for the world-changing disease. Things were moving at a fast pace. Every day, countless papers were published with clues to how this virus invades our cells and wreaks havoc in our bodies, with the hope of slowing its spread and helping the infected.
Our team at the University of Copenhagen’s Novo Nordisk Foundation Center for Protein Research wanted to contribute to the global research effort by studying the interactions between viral and host proteins. Specifically, we wanted to look for the role of a type of protein interaction that is relatively new to science, one that involves short stretches of amino acids known as SLiMs (short linear motifs). While the traditional structure-begets-function mentality regarding protein interactions had largely overlooked the unstructured protein regions and the short motifs within them, the past few decades have challenged this view, as researchers have documented the importance of SLiMs in a widening range of cellular functions.
What first drew our attention to SLiMs at the onset of the COVID-19 pandemic was the increasing recognition that many pathogens have evolved to “mimic” host SLiMs. With only a few amino acid changes, viruses can become equipped to interact with key host proteins and thereby meddle in critical cellular functions to promote viral proliferation or avoid detection by the immune system. In fact, many SLiMs were first discovered in viruses before they were described in non-viral proteins.
Together with our collaborators in Sweden and London, we developed a novel pipeline to discover SLiMs in viral-host interactions. Our work revealed a specific interaction between SARS-CoV-2’s nucleocapsid (N) protein, which packages the viral RNA and is critical for viral replication, and the human stress granule proteins G3BP1 and G3BP2. A cell forms stress granules upon sensing a stressor such as a virus. Alongside other functions, this causes the cell to stop protein production and minimize energy use, which can hamper viral functions, and viruses have evolved to disrupt this and other defense mechanisms.
In the late 1990s, it became clear that unstructured protein regions were not only abundant, they mediated important cellular functions.
In this context, we showed that SARS-CoV-2’s N protein disrupts stress granules by binding to G3BP proteins via a SLiM-based interaction that supports viral replication. Furthermore, to exploit this SLiM-based interaction, we developed a strong peptide inhibitor that significantly hampered viral proliferation in cells. This exposes an attractive avenue to follow for antiviral drug development, focusing on these short and elusive motifs, shown to be critical components in viral-host interactions.
As we look back at the evolving field of protein interactions, it’s exciting to consider just how recently the discovery of SLiMs rests on the timeline. Protein regions that were once ignored, grouped with others that lacked a defined structure, are now a growing focus in protein research, providing a new lens through which to study cellular function and disease. Beyond the more traditional view that protein interactions are mainly mediated by well-structured, three-dimensional domains whose interactions tend to be strong and long-lasting, we are coming to appreciate the transient and complex nature of proteins and the signaling networks they build, the foundation of cellular biology as we know it.
SLiM BASICSIn the early days of studying protein behavior, researchers recognized that large, structured protein domains often interacted with each other in a lock-and-key fashion, fitting together almost like puzzle pieces. Toward the end of the 20th century, however, the growing discovery of previously overlooked unstructured regions suggested that there was more to the story. One type of protein interaction mediated by the unstructured proteome involves short linear motifs (SLiMs), abundant stretches of up to 10 amino acids. Their interactions with other proteins are generally transient and weak, but SLiMs are nevertheless significant contributors to protein function and regulatory mechanisms in the cell. | |
Structured protein domains | SLiMs |
Approximately 50–200 amino acids, with several points of contact | Just ~2–10 amino acids, with only 2–3 that act as core binding determinants |
Distinct three-dimensional structure | Lack a three-dimensional structure |
Strong and often long-lasting interactions, such as in protein complex formation | Weak, transient interactions |
Bind domains of other protein partners, interactions that often resemble a | Typically bind to a conserved pocket on a globular protein domain |
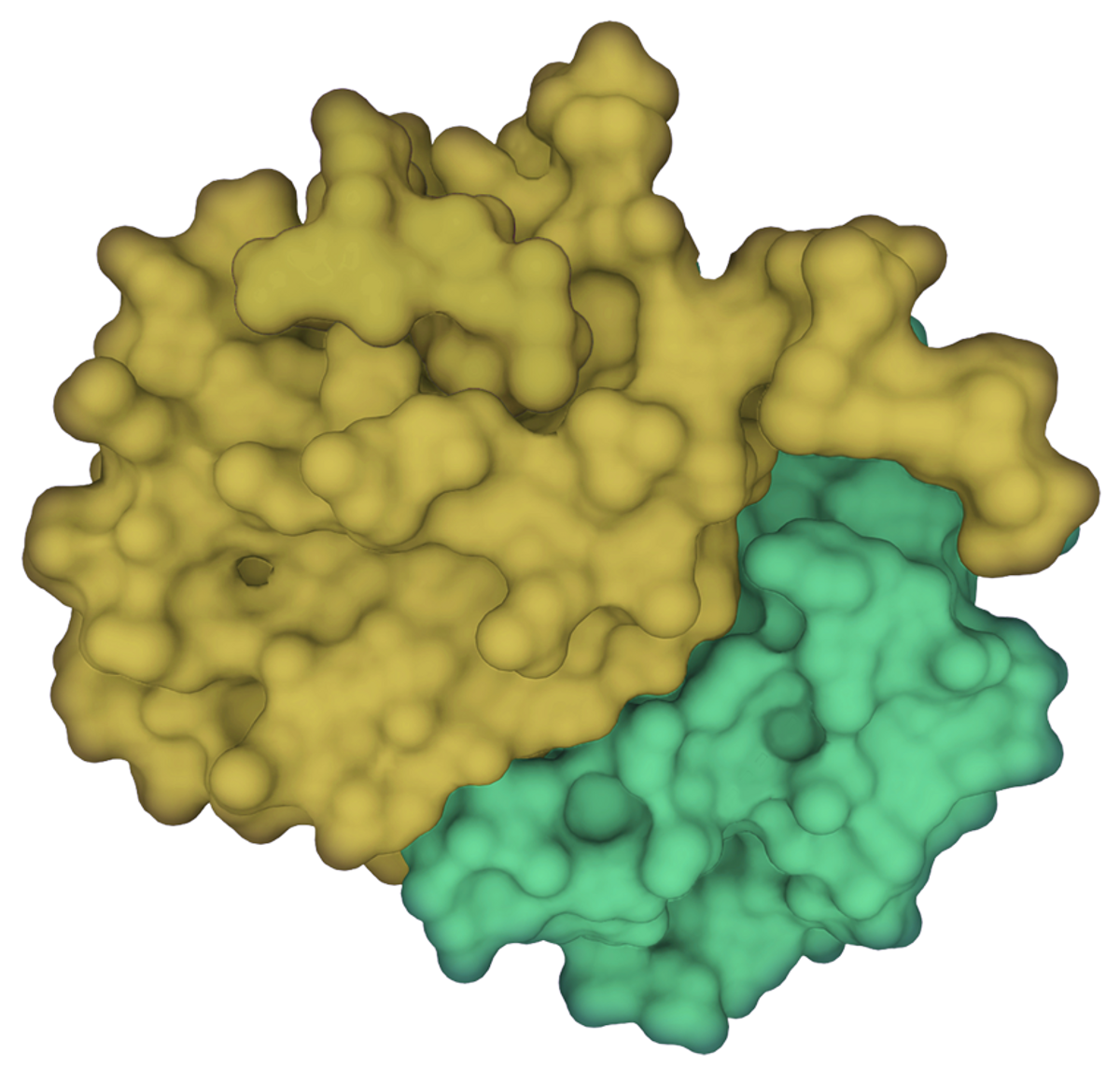 © Scott Leighton | 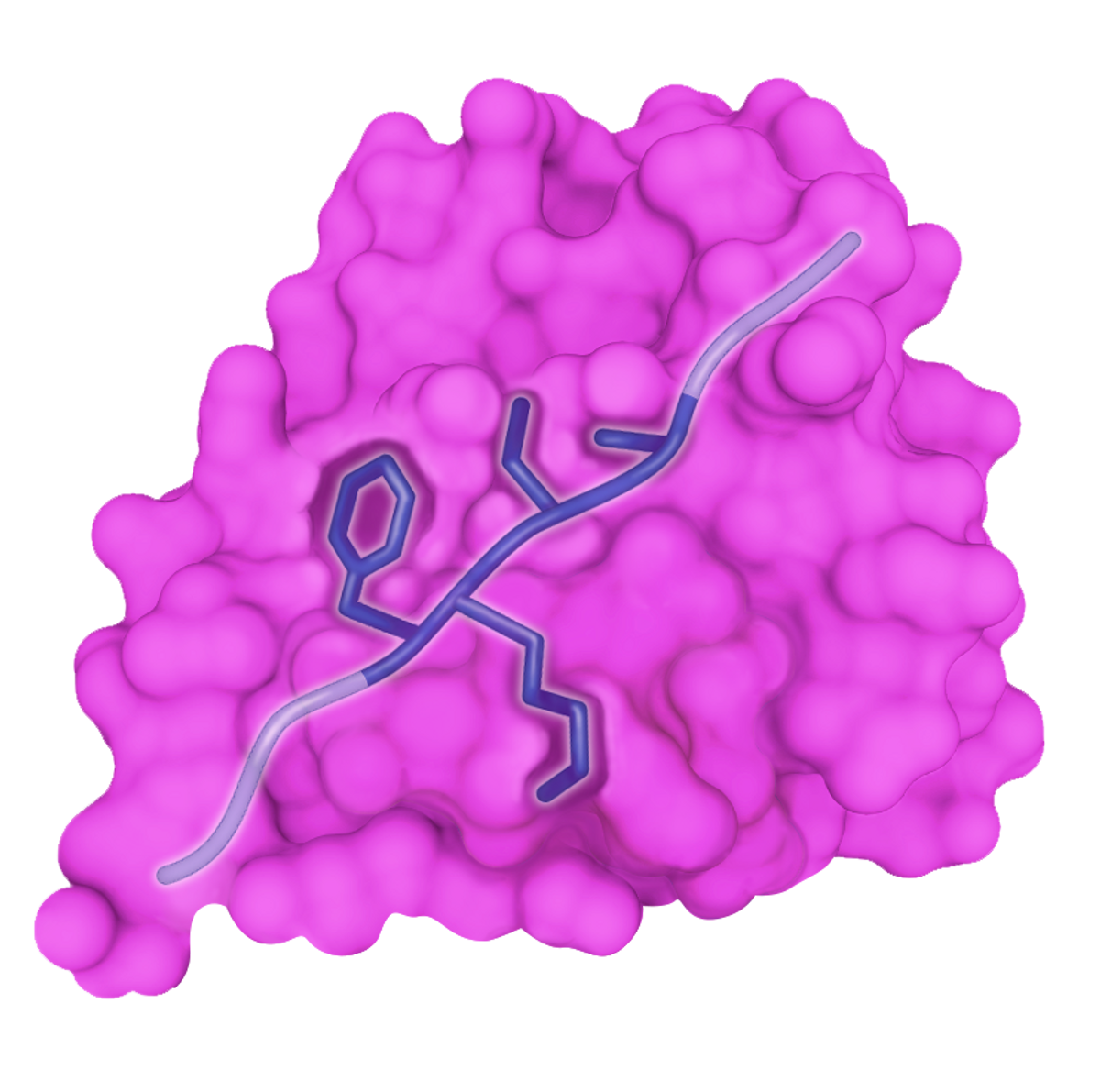 © Scott Leighton |
Paradigm shift
There are more than 20,000 proteins encoded in the human genome, and if you look at any structure in the RCSB Protein Data Bank, you will likely find that one part is consistently missing—a region that researchers could not solve. This is commonly seen in protein regions that lack a three-dimensional structure. For decades, these cases were dismissed as exceptions because the scientific community thought that a protein’s distinct three-dimensional structure determined its function, governing how it can interact with other proteins, like a key fitting neatly into a lock.
This idea was first described by German chemist Emil Fischer at the turn of the 20th century, and his hypotheses largely aligned with the results of many researchers interrogating protein structure over the second half of the 20th century, determined with the help of novel techniques such as X-ray crystallography and nuclear magnetic resonance spectroscopy. But while these often strong interactions between well-structured protein regions undoubtedly hold a critical place in biology, the focus on them overlooked countless unstructured regions that exist in the proteome. That all changed in the late 1990s when it became clear that these unstructured protein regions were not only abundant, they mediated important cellular functions.
This shift in our thinking, coupled with new biochemical and computational tools, resulted in the recognition of a new class of proteins that are unstructured, or intrinsically disordered. But these posed a new challenge to protein biologists: How can we study such unstructured proteins if there is no structure to solve? Where do we start?
Turning to bioinformatics, researchers uncovered many interactions within disordered protein regions, with the majority of interactions taking place between short stretches of just 2 to 10 amino acids—SLiMs, a term coined in 2006 by a group at the University College Dublin Conway Institute of Biomolecular and Biomedical Sciences. Compared with other types of interactions, these motifs bind weakly and transiently, often to conserved pockets on globular proteins. (See illustration on page 14.)
In the ’90s and into the early 2000s, investigations into SLiMs linked these motifs to many important cellular functions in higher eukaryotes, including protein localization, gene expression, cell cycle control, and protein degradation via the proteasome. SLiMs work alongside structured protein domains to ensure the maintenance of cellular signaling networks. Given their importance, it isn’t surprising that SLiMs are often dysregulated in diseases such as cancer. Ongoing computational efforts aid in motif discovery and help to grow databases compiling unstructured proteins and their mounting numbers of known interactions. A recent prediction estimates that the number of SLiMs in the human proteome exceeds 100,000, and that number skyrockets to nearly a million when post-translational modifications are considered.
Because they are so short, SLiMs can easily arise through mutation. They thus offer a versatility that structured protein domains cannot achieve alone, providing a fast and simple basis for new protein interactions and thereby allow organisms to rapidly adapt to their changing environment. This feature, however, also makes these motifs easy targets for pathogens to tap into host biology.
SLiMS IN SARS-COV-2 INFECTIONSARS-CoV-2 appears to take advantage of the host stress granule machinery deployed in response to viral infection to favor its proliferation. Specifically, our work shows that its nucleocapsid (N) protein, responsible for encapsulating viral RNA and coordinating replication and other functions, contains a SLiM that competes with cellular proteins in binding with stress granule–forming proteins, namely G3BP1 and G3BP2 (G3BPs). In doing so, the virus effectively promotes its proliferation while dampening the cell’s antiviral defenses. Targeting these SLiM-mediated protein interactions may one day prove to be a feasible antiviral therapy approach. |
Normal stress responseIn response to viral infection or other stressors, cells form stress granules, membraneless organelles that contain host mRNA, viral RNA, translation factors, and RNA-binding proteins. These include G3BPs, which bind to other stress granule proteins via an ΦxFG SLiM, where Φ represents a hydrophobic amino acid, x represents any residue, and F and G represent phenylalanine and glycine, respectively. This response causes cells to limit their energy use and restrict protein production. Because viruses need the cellular machinery to produce their viral proteins and proliferate, the stress response can hamper this, with stress granules often associated with an antiviral role. 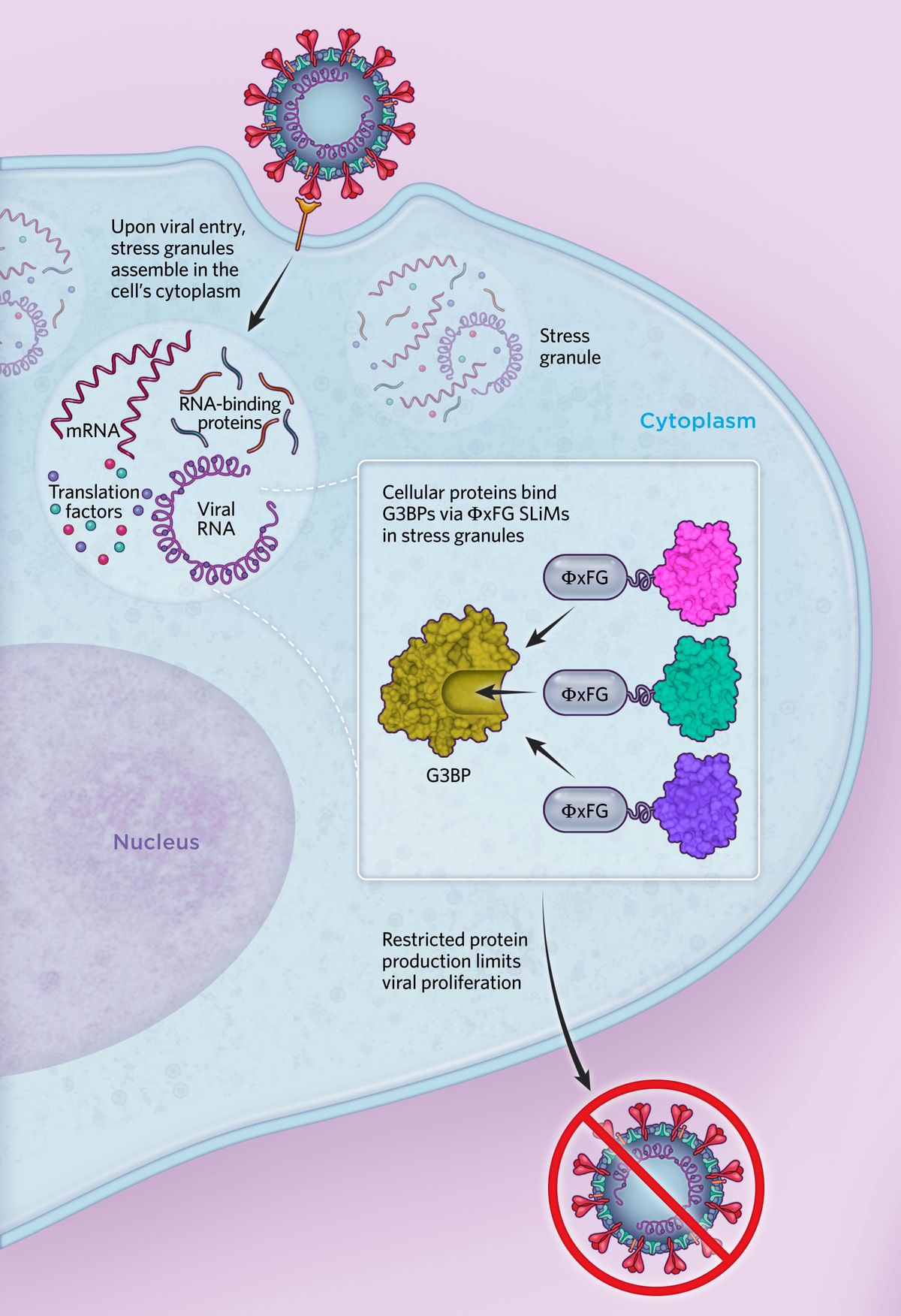 © Scott Leighton |
SARS-CoV-2 infectionIn the case of SARS-CoV-2, we found in human cells that the N protein is able to bind G3BP proteins via an ΦxFG SLiM, possibly to localize viral replication to stress granules in early infection. As levels of the SARS-CoV-2 N protein increase in later infection stages, N effectively displaces all cellular proteins from G3BPs and disrupts cytoplasmic stress granules. This ultimately promotes viral proliferation and the dampening of antiviral defense mechanisms. 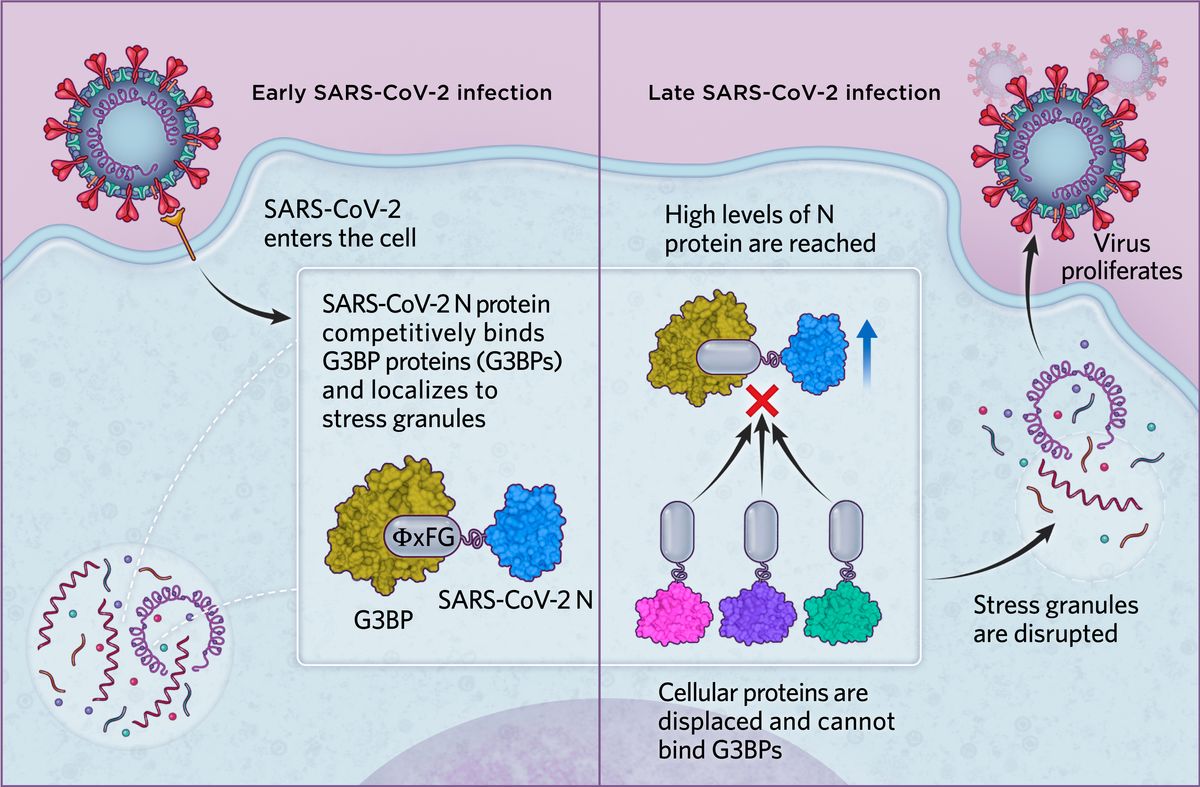 © Scott Leighton |
Targeting stress granule formationBased on these findings, we designed a peptide inhibitor containing ΦxFG-like SLiMs that bound strongly to G3BPs, preventing the binding of SARS-CoV-2 N protein in human cells. This peptide (G3BPi) inhibited viral proliferation in a monkey cell line commonly used in SARS-CoV-2 studies, though the specific downstream mechanisms remain to be elucidated. 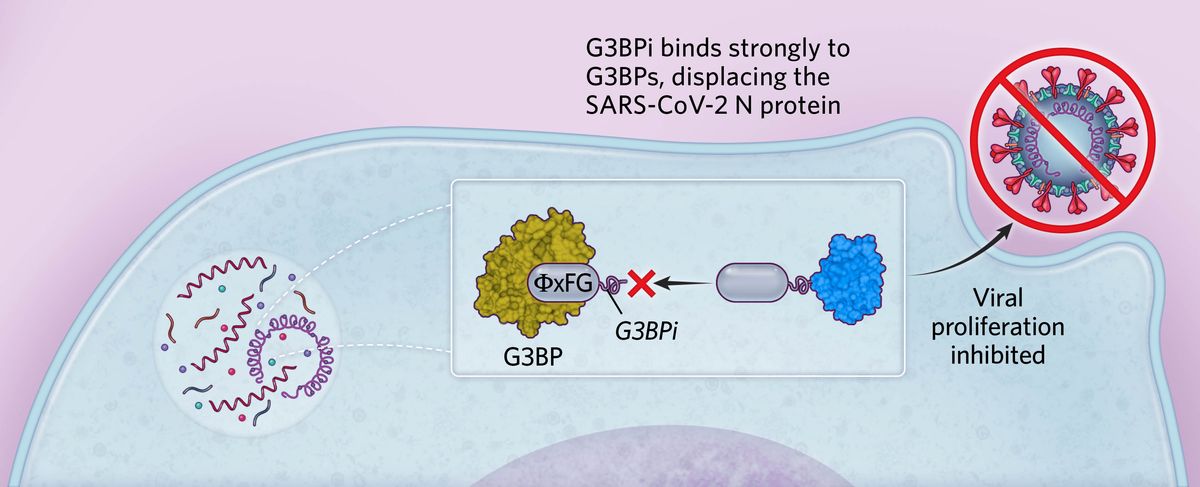 © Scott Leighton |
Viral hijacking
Viral protein sequences that mimic cellular SLiMs interfere with a range of host processes, exploiting cellular transport machinery, manipulating signaling pathways, and otherwise making themselves at home. In 2018, for example, our group showed that the nucleoprotein of Ebola virus mimics a SLiM binding a host phosphatase to facilitate viral transcription. And the LMP1 protein from the Epstein-Barr virus contains a SLiM that mimics a motif in CD40—a protein involved in the activation of antigen-presenting cells—allowing the virus to interfere with the host’s immune defense.
Several years ago, our Uppsala University colleague Ylva Ivarsson and her collaborators showed that proteomic peptide-phage display (ProP-PD), a technique useful for screening other protein interactions, can help identify SLiM-based protein interactions. In a nutshell, this approach involves a library of bacteriophages that are engineered to display peptides in their exposed coat protein. The phages are then presented to bait proteins, some of which will bind to the displayed peptides. Bound phages are then sequenced to find out which peptide motifs interacted with which bait proteins. To apply this to SLiMs, researchers engineered phages to present unstructured protein regions.
Using this technology, we and our collaborators designed a unique ProP-PD library, with phages displaying nearly 20,000 unstructured regions of more than a thousand viral proteins from 229 RNA viruses, including 23 coronaviruses. We presented these to known globular domains from human proteins that had been previously linked to viral-host interactions, uncovering a total of 117 SLiM-based human-coronavirus protein interactions. Turning our focus to SARS-CoV-2, we found that a peptide containing an unstructured region of the viral N protein was binding a specific domain in the human proteins G3BP1 and G3BP2.
G3BPs have been linked to innate immune signaling and are known effectors in the assembly of stress granules, dynamic structures composed of proteins and RNA that form in response to diverse cellular stresses such as viral infections. During the stress granule response, cells will limit their energy expenditure and, among other functions, restrict protein production. Because viruses need cellular machinery to produce their viral proteins and proliferate, stress granules can hamper this co-option. But viruses have evolved a counterattack.
Various studies have described multiple viral proteins that can recruit G3BP1/2 and other stress granule components to disrupt stress granule formation or facilitate viral replication. More-recent work has revealed that the SARS-CoV-2 N protein interacts with the G3BP proteins and that this leads to the disruption of stress granules. These studies even pointed to the N protein’s amino-terminal domain, where we found the G3BP-interacting SLiM, as being central in this role. We thus suspected that this SLiM—a so-called ΦxFG SLiM, where Φ represents a hydrophobic amino acid, x represents any residue, and F and G represent phenylalanine and glycine, respectively—could help promote viral replication in the face of a cellular stress response. (See illustration on page 14.)
SLiMs offer a versatility that structured protein domains cannot achieve alone, providing a fast and simple way for new protein interactions to arise.
We transfected cultured human cells with the SARS-CoV-2 N protein, either containing the ΦxFG motif or a mutated motif, and used live cell microscopy to image G3BP1 and thereby visualize stress granule formation in response to an introduced stressor, the compound arsenite. We found that stress granules were disrupted in cells expressing the N protein with an intact ΦxFG but not with the SLiM-mutated N protein. We then infected monkey cells (commonly used in studies of SARS-CoV-2 and other viruses) with SARS-CoV-2 and examined the localization of G3BP1, the N protein, and viral RNA using immunofluorescence. Intriguingly, when levels of SARS-CoV-2 N were relatively low, we saw normal stress granule formation, with all three molecules found clustered in the granules. But as infection advanced and viral N protein levels increased, we could no longer detect any G3BP1 granules. This made us suspect that the viral N protein is tapping into the stress granule machinery at early stages of infection, but once a threshold level of N viral protein is reached, the stress granules are disrupted.
Still, the mechanism by which this all happens wasn’t clear. We wondered which cellular proteins might be using this SLiM to bind the G3BPs—that is, which cellular proteins was the viral SLiM mimicking? If we could learn more about the process in the absence of infection, perhaps we could obtain a clue about the ΦxFG-mediated pathways the virus was tampering with.
For this we prepared a second ProP-PD library, this time displaying disordered protein regions of the human proteome, and used the conserved SLiM binding domain on G3BPs as bait. We identified 72 peptides that bound these proteins, with most of them containing an ΦxFG SLiM. Nineteen of these belonged to stress-granule associated proteins. Because the N protein is the most abundant viral protein during infection, we hypothesized that it could compete with these host proteins for binding to the G3BP proteins. Sure enough, quantitative mass spectrometry revealed this to be the case, with binding analyses confirming that ΦxFG-containing host proteins are displaced by the SARS-CoV-2 N peptide.
We considered whether these ΦxFG-mediated interactions could be targeted for therapeutic applications and sought a high-affinity peptide that would prevent viral N protein–G3BP interactions. Aided by previous research on G3BP binding to viral proteins in another RNA virus, we engineered a G3BP inhibitor (G3BPi). The designed peptide inhibitor successfully prevented the N protein binding to G3BP1 in cultured human cells and potently inhibited SARS-CoV-2 proliferation after 16 hours of infection. Excitingly, earlier this year a research group at the department of biochemistry at the University of California, Riverside, solved the structure of G3BP1 bound to SARS-CoV-2 N, confirming that this molecular interaction is largely mediated by the ΦxFG SLiM. The results revealed that the SLiM occupies a conserved pocket in G3BP1’s globular domain, validating our conclusions and shedding light on our proposed binding mechanism. Furthermore, it serves as a key example of a SLiM binding a conserved pocket on a globular protein domain, elucidating the nature of SLiM-based interactions and highlighting their therapeutic potential.
SLiMs are shaping up to be major regulatory motifs in cellular biology, from stress granule formation to innate immune signaling. In the context of infection, this raises questions about how a pathogen can rewire host protein networks to its own benefit. Despite their abundance and functional importance, super-short and quickly-evolving SLiMs are challenging to study. Traditional biochemical methods and technologies such as mass spectrometry screens tend to be biased toward stronger binding interactions. And although computation can allow us to predict the existence of SLiMs, experimental validation is still needed.
These are exciting times, as we move forward holding a flashlight in what seems to be a dark room filled with an overwhelming number of protein interactions that support life at the most basic level. But with novel approaches such as ProP-PD, alongside up-and-coming biochemical and computational tools, the field is becoming equipped to fully explore the unstructured proteome. Results from our group and others remind us to keep an open mind regarding how proteins function, as it’s now abundantly clear that structure cannot tell us the whole story.