ABOVE: Before modern imaging technology, Makio Murayama constructed large models to relate a molecule’s structure to its potential function. Gerry Hecht, Office of NIH History and Stetten Museum
Today, the cause of sickle cell disease is well understood to be a point mutation in a hemoglobin gene called HBB that makes red blood cells grow stiff and misshapen. Sixty years ago, however, top scientific minds struggled to understand the mechanisms of that so-called sickling of cells. The answer came, in part, thanks to a massive physical model of the protein deoxyhemoglobin (hemoglobin not yet bonded to oxygen) that was meticulously assembled over six years by Japanese-American chemist Makio Murayama.
Born in San Francisco in 1912, Murayama had earned master’s degrees in biochemistry and physics by 1940. Then came World War II, during which Murayama’s family was sent to an internment camp in Idaho, while Murayama himself traveled to Chicago to work on what eventually became the Manhattan Project. He never started, however: he was turned away and surveilled by the FBI due to his heritage and instead studied treatments for blood diseases at Children’s Hospital of Michigan.
A subsequent PhD from the University of Michigan led Murayama to a postdoc at Caltech working with Linus Pauling, who had famously discovered that sickle cell disease was hereditary. In 1958, Murayama moved to the National Institutes of Health (NIH), where he spent years as the only full-time scientist pursuing basic sickle cell research. By day, Murayama conducted lab experiments to understand the mechanisms of sickling. By night, he took another approach: constructing a 70,000-piece model of deoxyhemoglobin in his basement to study how the molecule’s structure influenced its role in sickle cell disease.
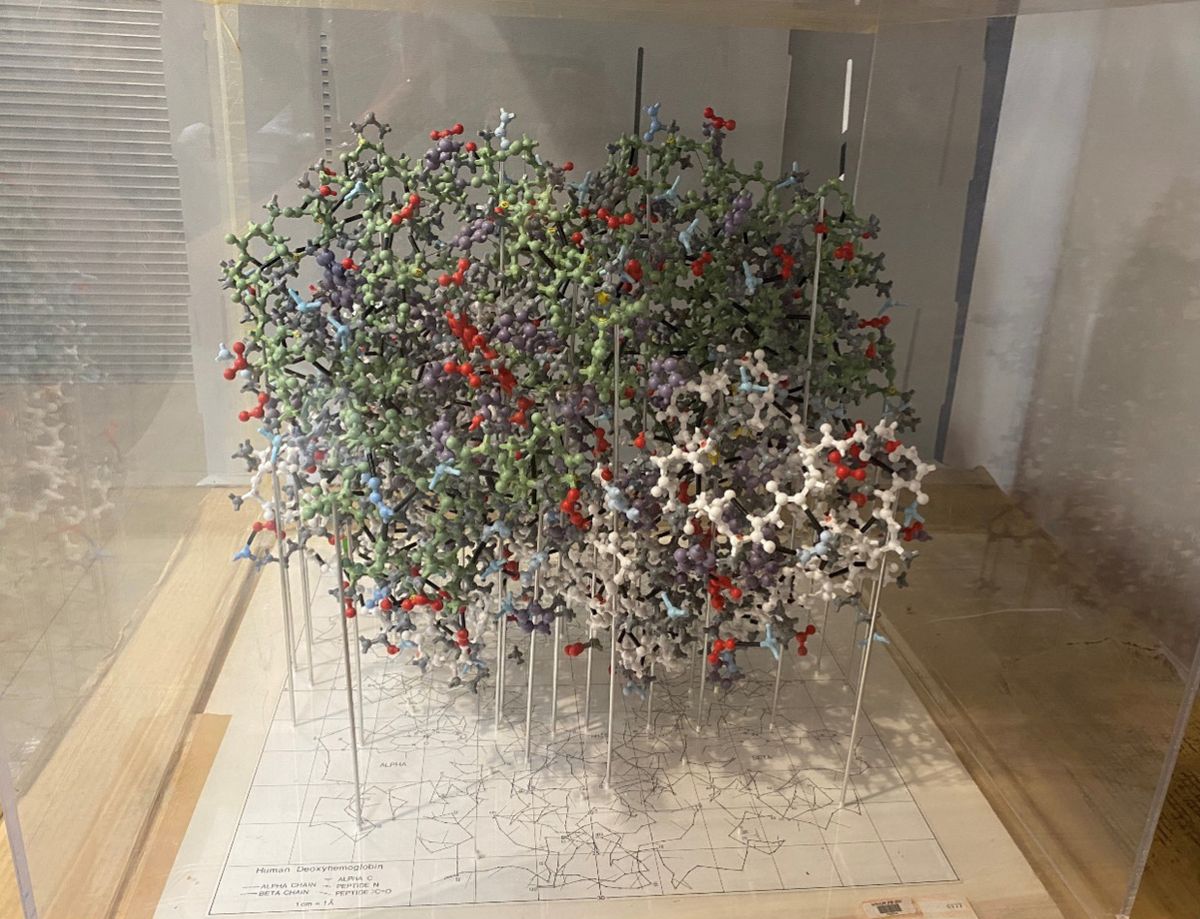
Alan Schechter, a senior scientist and historical consultant at the National Institute of Diabetes and Digestive and Kidney Diseases who knew Murayama personally, tells The Scientist that the model ended up being inaccurate in key ways (it originally had an internal structure that didn’t exist in the real protein). But it was good enough to help Murayama discover the mechanism of sickling, in which faulty hemoglobin produced by mutant HBB polymerizes into stiff fibers that give sickle blood cells their rigid, hooked shape. Schechter says that Murayama “was ahead of his time” in connecting the structure of a molecule to its function, even if he did so imperfectly.
Murayama later developed a laboratory test to detect sickle hemoglobin by triggering temperature-mediated, reversible polymerization, and he was instrumental in the discovery that urea can break the bonds between sickle hemoglobin molecules, according to a 1972 paper by his close collaborator, Robert Nalbandian. Murayama advocated urea’s use as a therapeutic for sickle cell disease, partnering with clinicians in Ghana to test urea— “still an experimental drug” then, Schechter notes—on patients in the 1970s. These studies would ultimately complicate his legacy, as urea, despite successfully reversing sickling, proved dangerous at the concentrations necessary for a therapeutic effect. Today, sickle cell is treated with some urea-based pharmaceuticals, but also by blood transfusions. A number of gene therapies are currently under development, too.
Murayama died in 2012, at age 99. Schechter says that even though Murayama made errors in his career, his work to “realize the importance of molecular structure . . . had a very important effect on [sickle cell] therapy.”