Stay up to date on the latest science with Brush Up Summaries.
What Is Spatial Biology?
Spatial biology is a series of techniques with which scientists collect detailed cellular information and examine the positional context of cells in a tissue. Individual cells differentially use their genes, RNA, and proteins across tissue types. Spatially resolved analyses provide insight into new strategies to prevent or treat diseases, including infection, cancer, neurological conditions, and metabolic disorders.1,2
What Can Researchers Learn from Spatial Biology?
Spatial transcriptomics
Transcriptomics encompasses studies in which researchers examine gene expression dynamics and heterogeneity with RNA sequencing. In spatial transcriptomics—also referred to as spatial genomics—methods for capturing positional transcriptome information often unite the previously separate realms of imaging and sequencing.2 Scientists rely on a variety of microscopy-based methods and transcript-complimentary probes to record the location of different RNA species in tissues, either directly or prior to targeted sequencing. Some examples of these methods include in situ hybridization (ISH), in situ sequencing (ISS), arrays of spatially barcoded probes paired with next-generation sequencing (NGS), and microdissection.1
Spatial proteomics
Spatial transcriptomics is not the only -omic approach to studying the immense complexity of biological systems. Just as in situ hybridization-based imaging and state of the art sequencing have brought spatially-resolved transcriptomes to the research forefront, scientists employ established and novel methods to spatially analyze protein distribution at the tissue, cell, and subcellular levels. Spatial proteomic methods include immunohistochemistry (IHC), immunofluorescence (IF), mass spectrometry (MS), and cytometry. These techniques have varied coverage depth and throughput.3
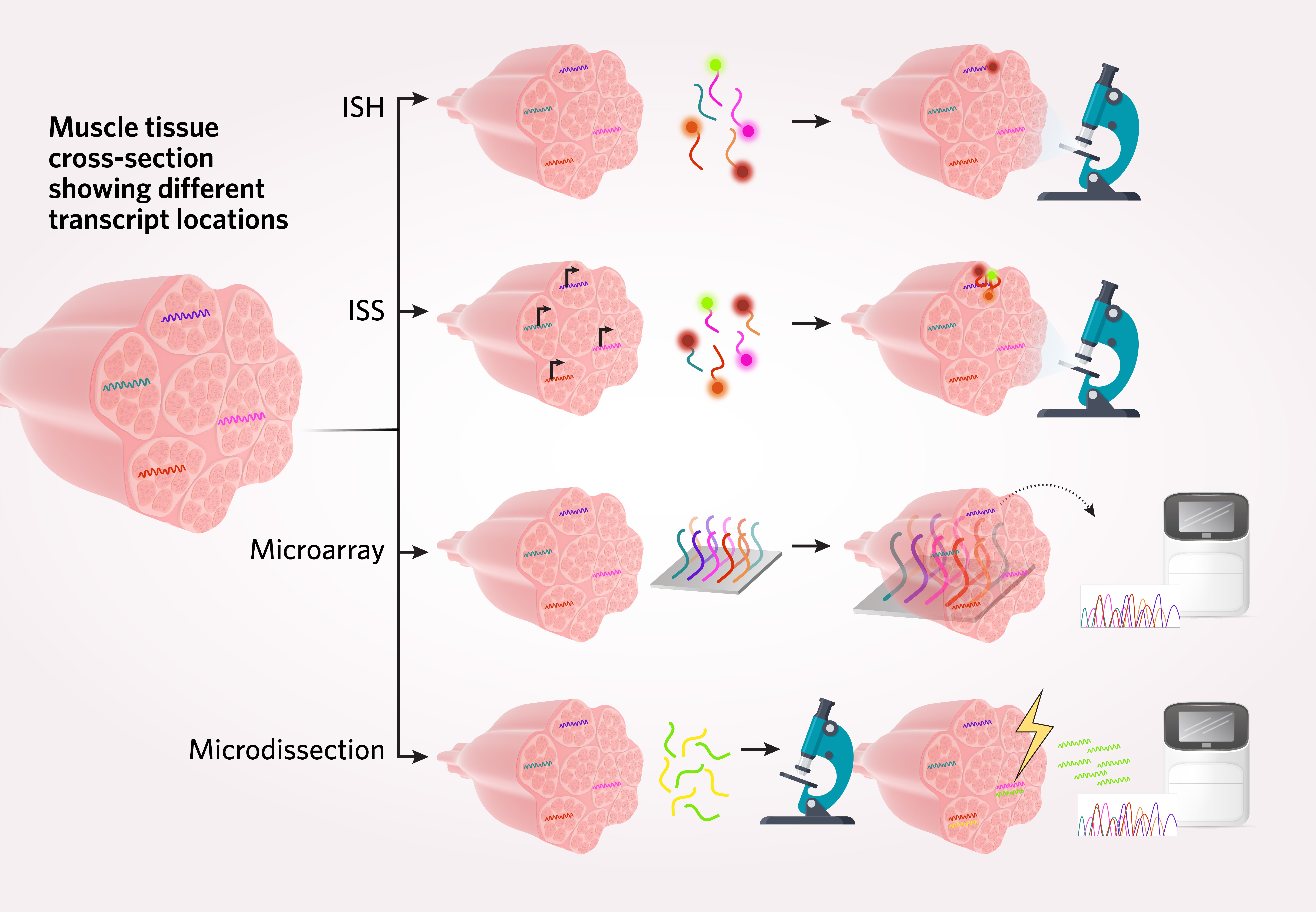
Advantages and limitations of current spatial biology methods
Before the advent of spatial transcriptomics, single-cell RNA sequencing methods largely required that scientists break tissue apart into individual cells (e.g., islet cells from pancreatic tissue). As a result, prior technologies lost the spatial context needed to fully understand biological processes such as cell-to-cell interactions between normal and diseased tissue, and how unique cell types contribute to heterogeneous tissues.
Spatial transcriptomics methods address this challenge, capturing entire areas of tissue. Using these methods, scientists infer single-cell resolution of the transcriptome with spatial context.3 However, spatial analysis of transcriptome-wide information of all single cells in a tissue sample is not yet a routine process.2 Some additional challenges to spatial genomic and spatial proteomic analyses include tissue suitability for specific methods (e.g., human brain tissue autofluoresces, which may complicate fluorescence imaging-based techniques) and detection limitations for rare cells and low copy number transcripts. In addition, many imaging and sorting methods rely on low-throughput, antibody-based detection, which can be a challenge for molecular targets without established antibodies.1,3
Spatial Biology in Practice: Mapping the Cell Atlas Across Tissue Types
Spatial biology is a fast-growing field, driven in part by improved accessibility of NGS, as well as initiatives such as the Human Cell Atlas (HCA).1 The HCA project is an international collaboration of scientists aiming to define all human cell types in terms of distinctive molecular profiles and cellular descriptions such as location and morphology.4 Researchers in the HCA community have already contributed new fundamental biological discoveries with potential for clinical applications.5
For example, HCA researchers have reported single cell data that highlights the cellular heterogeneity of a multitude of tissues, including the heart, liver, intestines, pancreas, thymus, and brain. Spatially examining transcriptomes of different tissues leads to new insights, such as an improved understanding of sex-related risk factors in heart disease. It also reveals complexity that was previously overlooked, such as the identification of epithelial progenitor cells in the liver or pancreatic cell type-specific neighborhoods with unexpected cell to cell interactions.5
Beyond novel insights into specific tissue types, the HCA project aims to generate a comprehensive reference list of all identities and characteristics of the cells throughout the body. Such a list will accelerate fundamental understanding and translational science. The power of spatial biology approaches in HCA research will inform scientists about which cells express different genes of interest, what cell types are present in each tissue, and which cell types co-occur in close spatial proximity to one another.5

References
- C.G. Williams et al., “An introduction to spatial transcriptomics for biomedical research,” Genome Med, 14(1):68, 2022.
- V. Marx, “Method of the Year: spatially resolved transcriptomics,” Nat Methods, 18(1):9-14, 2021.
- A. Mund et al., “Unbiased spatial proteomics with single-cell resolution in tissues,” Mol Cell, 82:2335-49, 2022.
- A. Regev et al., “The Human Cell Atlas,” eLife, 6:e27041, 2017.
- R.G.H. Lindeboom et al., “Towards a Human Cell Atlas: taking notes from the past,” Trends Genet, 37(7):625-30, 2021.