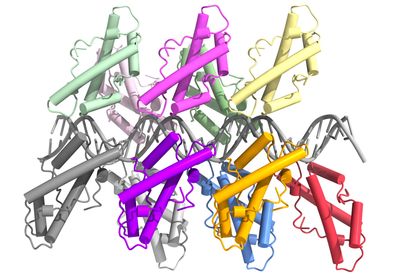
ABOVE: The bacterial histone Bd0055 SHAWN LAURSEN AND KAROLIN LUGER
Human cells manage to store approximately two meters of DNA in a nucleus one-tenth the width of a human hair. This feat relies on a family of proteins known as histones, which serve as spools around which DNA can wrap itself. In addition to creating a compact genome, histones play essential roles in gene regulation, genome replication, and cell division—in eukaryotes, anyway.
With the exception of two known proteins, bacteria are generally thought to lack histones, having evolved an independent set of DNA-folding histonelike proteins instead. As evolutionary biologist Tobias Warnecke at the LMS London Institute of Medical Sciences explains, bacterial DNA-packaging proteins have 3D structures that are “clearly distinct.” Still, the general absence of histones in bacteria is puzzling given that bacteria exchange genes with archaea and eukaryotes—a process called horizontal gene transfer—and could have adopted more histones in this way.
See “Conserved Chromatin?”
Now, Warnecke and colleagues are challenging the idea that these space-saving proteins are not commonplace in bacteria. In a preprint uploaded to bioRxiv this January that has yet to undergo peer review, the team describes hundreds of potential histone proteins in bacteria, including one that they predict interacts with DNA in an unusual way.
In an email to The Scientist, Remus Dame at Leiden University, Netherlands, who was not involved in the study, says “it is a very interesting study as histone[s] are essentially undescribed in bacteria.”
Warnecke wasn’t the first to look for histone in prokaryotes, but such searches, which tend to use eukaryotic histones as references, have only yielded a few potential candidates. That may be because bacterial histones are smaller, Warnecke hypothesized. So, with the help of Antoine Hocher, a postdoctoral researcher in his lab, Warnecke set out to determine if bacterial histones have much shorter sequences than eukaryotic ones.
Hocher sifted through over 18,000 bacterial genomes using shorter histone-related proteins from archaea as references. With this strategy, he discovered over 400 bacterial proteins that contain a signature 3D histone structure called a histone fold, suggesting histones could be scattered across many bacterial species.
With so many candidates to choose from, the researchers homed in on a histone in the species Bdellovirbio bacteriovorus, a bacterium that preys upon other bacteria. The team thought it might be a good candidate for finding histones because the bacterium changes size throughout its lifecycle and thus would need to carefully time DNA compression and unwinding. As a small predatory cell, it invades a prey bacterium, expands in size, and divides into multiple small predators, continuing the cycle.
The team detected a histone fold in an uncharacterized B. bacteriovorus’ protein called Bd0055 and wondered if this protein could help orchestrate DNA packaging. By observing that, in the presence of the protein, DNA moved much slower through pores in a gel, they confirmed that Bd0055 binds DNA. Deleting the gene turned out to be lethal in B. bacteriovorus cells both inside and outside prey bacteria, indicating that the protein is essential throughout the lifecycle.
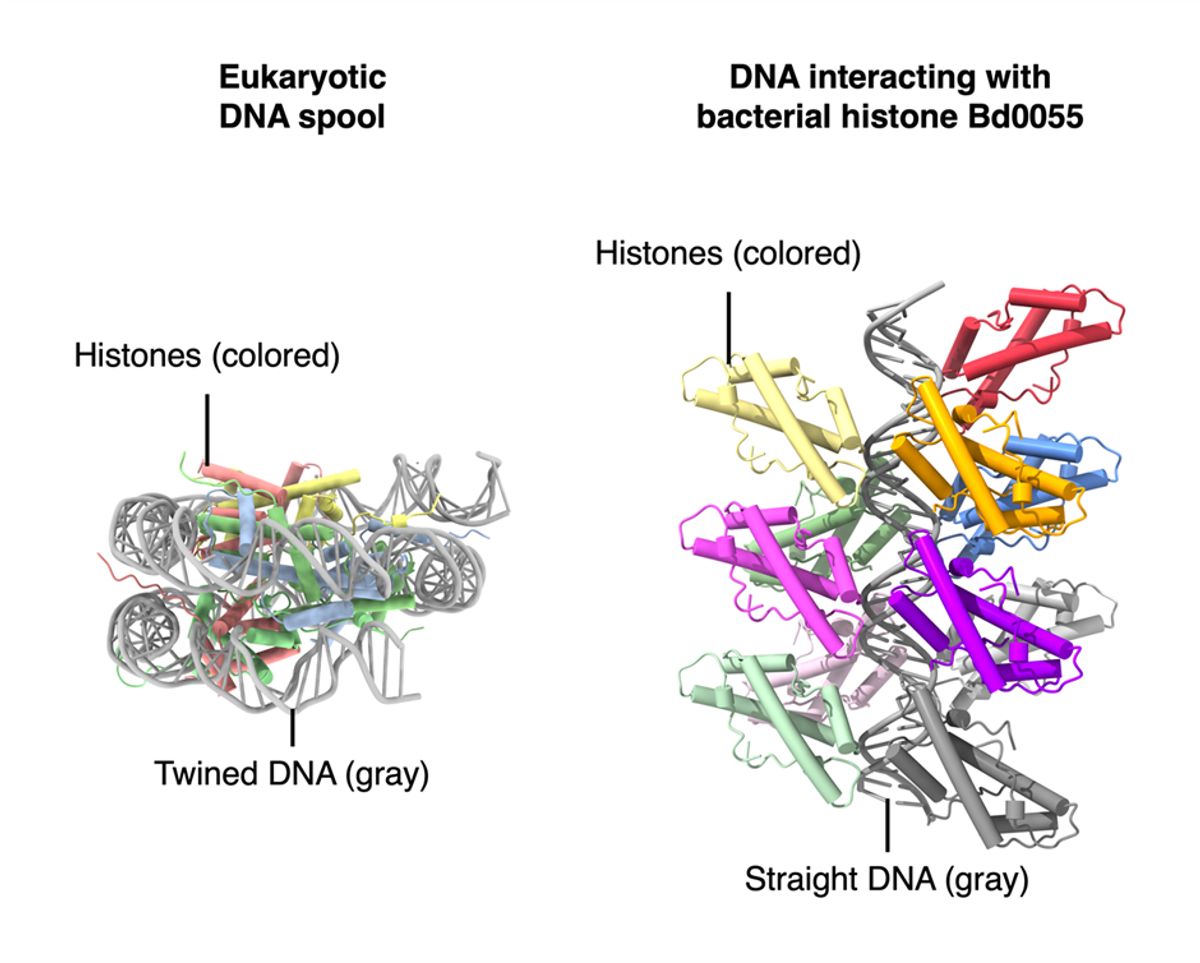
With all evidence thus far suggesting Bd0055 could be a typical histone protein, the team set out to determine if Bd0055 forms spools for DNA to wrap around. Computer simulations carried out by Shawn Laursen at the University of Colorado Boulder revealed that the protein-protein interactions that are needed to form spools would be unstable for this protein. Instead, the simulations revealed that Bd0055 might bind to the outer edges of DNA strands. Warnecke has never seen other histones behave this way. “There’s really nothing like the edge-on binding,” he says.
Rather than packaging the DNA to occupy less space, the authors argue that the edge-on binding could straighten the strands and protect the DNA from over-compression during division into smaller cells. Indeed, straightened DNA spirals have been observed in this bacterium. Nessa Carey, a molecular biologist who works on histones and was not involved with the study, agrees, noting that “it’s a possible interpretation” but also wonders if the protein may have a shielding action against bacteriophages or arsenal in the prey bacterium. The protein could be “making DNA harder to get at and attack.”
While Dame is intrigued by the findings, he notes that the function of the protein needs to be confirmed inside cells. Carey says that it’s “incredibly thorough structural work” but mentions that the lethal outcome of deleting the histone is unfortunate: “If all you get is a lethal phenotype, it tells you it’s important, but it doesn’t tell you why.” She proposes that more sophisticated genetic variants will be needed to build on these findings.
B. bacteriovorus is known to have exchanged genes with a distant relative, Leptospira interrogans, in the past, and the researchers found a similar histone protein in this species. Laursen says he plans to investigate this histone next. His supervisor, Karolin Luger, tells The Scientist in an email that because Leptospira histones share many features with Bd0055, the team suspects they might bind DNA “in the same unorthodox manner,” but there are also “intriguing differences.”
“We currently have a ‘beer bet’ in the lab on the outcome,” Luger says. “At any rate, it will be exciting to see whether this DNA binding mode is a general phenomenon for bacterial histones.”
Clarification (February 2): Dame’s comments have been amended to clarify that the protein’s function in cells is what needs to be confirmed, instead of its specific mode of DNA binding.