Stay up to date on the latest science with Brush Up Summaries.
What is Proteomics?
In an organism, thousands of proteins interact in a cell-type and tissue-specific manner to govern cell fates and functions. Protein-coding genes create tens to thousands of copies of different peptides in a cell. Further, protein expression varies from cell to cell and their levels change over time. Many of these proteins interact with each other, localize to distinct subcellular compartments, and undergo post-translational modifications, including phosphorylation, glycosylation, and ubiquitination. Investigating protein levels, composition, interactions, and structures within the cellular context is at the heart of understanding biological systems in health and disease.1-3
Proteomics is the large-scale study of proteins present at cellular and systemic levels. By generating comprehensive protein datasets, scientists understand the ebb and flow of protein expression in a tissue, how it differs from cell to cell, and how these differences illustrate the inner workings of an organism.1-3
What Does the Field of Proteomics Investigate?
Scientists employ three main approaches in proteomics studies: expression, structural, and functional proteomics.2,4
Expression proteomics determines where and when proteins are expressed and measures their quantities. This qualitative and quantitative approach can compare protein expression across conditions, such as health versus disease states, allowing researchers to identify disease-specific proteins. With this expression data, scientists can also identify new cell type markers, which allows them to label and manipulate their samples more accurately.2
Structural proteomics elucidates the three-dimensional structures of different proteins and their interactions with cell compartments, such as membranes, organelles, and nucleosomes.2
Functional proteomics allows researchers to examine protein functions and networks within a cell. By mapping the interaction of a specific protein with numerous partners, including unknown proteins, researchers predict how these interactions drive specific molecular and cellular pathways.2
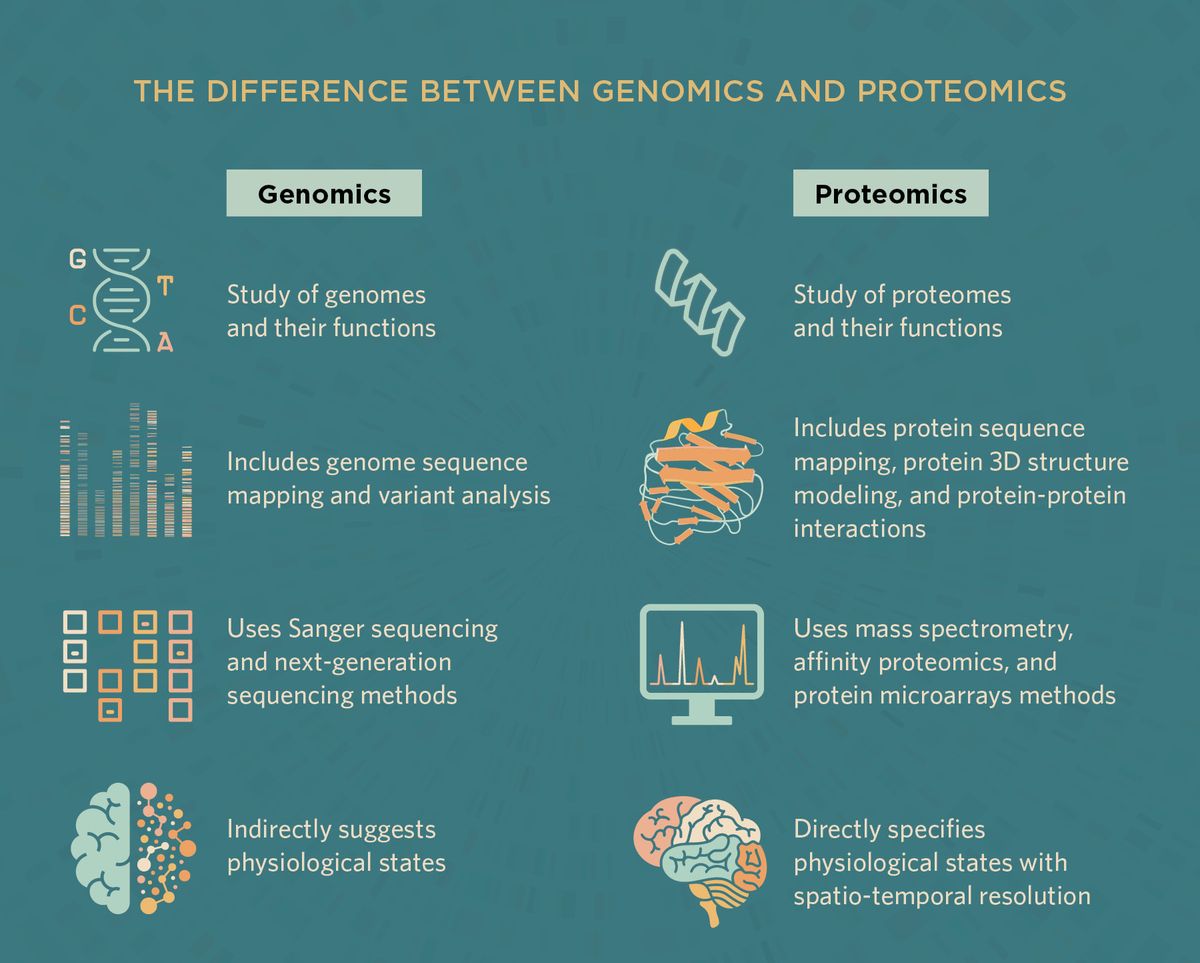
What Can Proteomics Reveal that Genomics Cannot?
Using genomics, researchers map exomes and whole genomes and identify genetic markers and gene variants. However, one key limitation of genomics is that the data only suggest indirect measurements of cellular states. Protein expression and regulation accurately reflect physiological states, which are measured with proteomics. Further, genomics data do not reveal protein levels, their dynamics across time, and post-translational modifications. With proteomics, scientists generate a map of the complex protein networks and their molecular interactions to gain direct insights into biological pathways.3,6
What Methods Do Scientists Use in High-Throughput Proteomic Experiments?
Mass Spectrometry
Mass spectrometry (MS)-based proteomics is the most comprehensive approach, allowing researchers to quantify protein levels and discover protein modifications and interactions. MS detects a peptide’s abundance by reading its fundamental properties, such as molecular mass and net charge. The mass gives information on protein identity, structure, and chemical modifications.7
-2.jpg alt=)
A mass spectrometer is equipped with a source, an analyzer, and a detector. The source uses gas or liquid ionization methods to produce charged peptide fragments. The analyzer separates these fragments based on their mass-to-charge ratios. Detectors enable signal detection and amplification in response to the charged species present in the analyzer.
MS-based proteomics is gaining popularity in all biological research fields, from discovery science in fundamental research laboratories to diagnostics applications, because of its analytic and quantitative power. However, mass spectrometry has several limitations, such as tedious protocol standardization and data analysis pipelines that demand time and expensive resources.
Affinity Proteomics
Affinity proteomics uses antibodies and other binding reagents, such as protein-specific detection probes, for proteome analysis. Affinity proteomics platforms offer high-throughput protein profiling, protein-protein interaction analysis, and post-translational modification detection from body fluids, cultured cells, and tissues. While highly efficient and robust, affinity proteomics is limited to well-characterized proteins with pre-existing antibodies and probes.8
In disease-specific applications, affinity proteomics allows for robust candidate biomarker quantification and facilitates new biomarker discovery and validation. For diagnostics, affinity proteomics is more advantageous than mass spectrometry because, in clinical applications, scientists want to profile multiple proteins at once in a short amount of time. For example, fast and cost-effective tumor biomarker identification is essential for early cancer detection and diagnosis.
Protein Chips/Protein Microarrays
Protein chips or microarrays facilitate large-scale, high-throughput proteomics where researchers can survey a cell’s entire proteome. These chips consist of probes on a support surface, such as glass, nitrocellulose membranes, or beads. Probes are ligands, chemical compounds, aptamers, or antibodies tethered to fluorescent dyes that selectively bind to proteins of interest present in the sample. Powerful laser scanners detect and quantify the fluorescent signal resulting from protein-probe interactions, where higher binding produces greater signal intensity.9
Because the workflows can be highly automated, protein chips allow for rapid and highly sensitive protein detection from small sample and reagent quantities. Researchers sometimes modify protein arrays to improve protein detection. For example, reverse-phased protein microarrays immobilize a set of proteins in the array to capture disease-specific biomarkers from an individual's sample.10
While protein microarrays capture numerous proteins at once compared to other proteomics methods, making sense of data and delineating protein concentrations and interactions from large-scale datasets remains a big challenge.9
How Do Researchers Analyze Proteomics Data?
A large-scale quantitative proteomics dataset is commonly represented as a 2D matrix of quantitative values for different peptides identified in a sample. Biostatistics and bioinformatics are required to interpret proteomics data.11,12
Regardless of the proteomics method used, scientists typically employ a similar data analysis workflow involving data standardization, protein annotation, and protein quantification. Depending on the method used for data acquisition, the results may include information about protein identity. For example, MS data include peptide spectra as mass-to-charge ratios that can be decoded with protein inference algorithms.5
At the discovery stage, researchers use algorithms and pipelines to identify their samples’ amino acid sequences, protein structures with putative binding pockets, and any post-translational modifications. Many bioinformatics algorithms also generate protein-protein interaction maps, allowing researchers to construct distinct biological pathways and determine how they associate with each other. Several of these methods use simulations to model biological networks. By computationally building complex cellular interactions, researchers get clues that help them design experiments to test proteins interactions and identify the consequences in vivo.11,12
Functional annotation of proteomics data determines a protein’s function by comparing databases that include biological pathway information. For example, gene ontology (GO)-based classification categorizes a gene or protein according to its functions, pathways, and structural domains.5,12 Using GO annotations, researchers can predict a protein’s molecular function along with which biological process they participate in a given cellular context.
References
- “What is proteomics?”, https://www.ebi.ac.uk/training/online/courses/proteomics-an-introduction/what-is-proteomics/, accessed on January 4, 2023.
- P.R. Graves, T.A. Haystead, “Molecular biologist's guide to proteomics,” Microbiol Mol Biol Rev, 66(1):39-63, 2002.
- S. Al-Amrani et al., “Proteomics: Concepts and applications in human medicine,” World J Biol Chem, 12(5):57-69, 2021.
- M. Cui et al., “High-throughput proteomics: a methodological mini-review,” Lab Invest, 102, 1170-81, 2022.
- C.M. Carnielli et al., “Functional annotation and biological interpretation of proteomics data,” Biochim Biophys Acta, 1854(1):46-54, 2015.
- “Genomics vs Proteomics- Definition and 10 Major Differences,” https://thebiologynotes.com/difference-between-genomics-and-proteomics/, accessed on January 4, 2023.
- A. Sinha, M. Mann, “A beginner’s guide to mass spectrometry–based proteomics, Biochem (Lond), 42(5): 64-9, 2020.
- M.D. Witte, “Modular approaches to synthesize activity- and affinity-based chemical probes, Front Chem, 9, 2021.
- C.S. Chen, H. Zhu, “Protein microarrays,” Biotechniques, 40(4):423-7, 2006.
- C. Paweletz et al., “Reverse phase protein microarrays which capture disease progression show activation of pro-survival pathways at the cancer invasion front,” Oncogene, 20, 1981-9, 2001.
- “Basic proteomics workflow,” https://idearesourceproteomics.org/wp-content/uploads/2017/09/Basic-Proteomic-Workflow.pdf, accessed on January 4, 2023.
- M. Turewicz et al., “BioInfra.Prot: A comprehensive proteomics workflow including data standardization, protein inference, expression analysis and data publication,” J Biotechnol, 261:116-25, 2017.