ABOVE: © ISTOCK.COM, BSD555
Like physical traits, behavioral traits have a heritable component, and personalities may resemble one parent’s more than the other. New research in mice finds that specific complex behaviors in these animals are shaped by genes inherited from just one parent.
Not only are alleles inherited from a mouse’s mom or dad expressed in unequal proportions in various cells in the brain and adrenal system—a phenomenon called genomic imprinting—but expressing the maternal or paternal allele leads to observable differences in the behavior and physiology of the offspring, according to a study published in Cell Reports on March 8. The scientists behind the research also found that maternal alleles shape the foraging behavior of male offspring, while the paternal alleles shape the behavior of female offspring. But why and how this happens is not yet clear.
See “Leaving an Imprint”
The paper results from nine years of research in which lead author Christopher Gregg, a neurobiologist and geneticist at the University of Utah School of Medicine, and other members of his lab developed a way of studying what he calls “behavioromics,” which involves letting mice go about their lives in a pseudo-natural setting and using a machine learning algorithm to screen for “hundreds of components of behavior in one assay, and discovering the phenotypic effects” of underlying genetic drivers, he explains.
For the new study, “I thought that was such a provocative idea to test,” Gregg tells The Scientist, “to determine whether mom and dad were affecting different decisions in the offspring.”
Exploring genomic imprinting
Gregg explains that he started working toward this project more than a decade ago when he was a postdoc in the lab of Catherine Dulac, a Harvard University molecular biologist. At the time, RNAseq—a technique used to determine and quantify the extent to which specific genes are being expressed—was new, and Gregg used an allele-specific version to reveal that for multiple genes, the allele from one parent is more likely to be expressed than the allele from the other, though the technology at the time couldn’t determine that this was a cell type-specific phenomenon. In 2010, he and Dulac published a pair of papers in Science that identified hundreds of genes that seemed to show preferential expression depending on the parent of origin, which is coded onto the DNA by allele-silencing epigenetic marks such as methylation or histone modifications.
See “Imprinting Diversity”
Gregg decided to zoom in and investigate whether some but not all cell types within a tissue may silence a parental allele, eventually demonstrating in a 2015 Cell Reports paper that this seems to be the case. It wasn’t yet possible to determine which parent’s alleles were being expressed in a given cell type, just that preferential expression of one parent’s allele over the other was occurring. The paper identified several brain regions—many located within the hypothalamus—that contained imprinted genes.
In the new paper, the researchers used a technique that allowed alleles from a mouse’s parents to each be stained in different colors. They then sectioned mouse brains and examined the fluorescent markers under a microscope, observing that allele silencing varied among individual cell types and clusters in specific regions. That reinforced the idea that the partial silencing observed in older studies came from averaging the effects of some but not all cells silencing a given allele.
Dulac, who wasn’t involved in the new study, says that “it’s a beautiful demonstration that in some circumstances, at least, this parental bias corresponds to full silencing of some alleles.”
See “Neural Circuit of Parental Behavior Mapped in Mice”
The study demonstrated that cells in 14 brain regions, including multiple parts of the hypothalamus, which helps coordinate the endocrine system, specifically express the maternal allele for the gene DOPA decarboxylase (Ddc)—which encodes an enzyme involved in making several neurotransmitters—and for a second gene called tyrosine hydroxylase (Th), which encodes another enzyme with a similar role. Meanwhile, the paternal copy of those alleles was preferentially expressed by cells in the adrenal gland, which produces hormones. These findings illustrate maternal and paternal control over the synthesis and distribution of hormones in offspring, at the cellular level, via the same genes, Gregg explains.
Whatever the mechanism is, it has to be pretty complex to explain these patterns.
—Robert Feil, Institute of Molecular Genetics, Montpellier-CNRS
“What’s unusual here is the diversity of the patterns that emerge,” says Robert Feil, a molecular geneticist who studies the mechanisms of imprinting at the Institute of Molecular Genetics of Montpellier-CNRS in France. What stood out most to Daan Noordermeer, a genomic imprinting researcher at the Institute for Integrative Biology of the Cell of University Paris-Saclay, was how the patterns of expression varied from the genes’ typical activity: usually, both parental alleles are expressed in the same cells, he tells The Scientist over email. Neither Feil nor Noordermeer was involved in the study.
Behavioromics: a big data approach to the epigenetics of behavior
Gregg says that the next step was to figure out what, if any, effects the imprinting patterns had on specific mouse behaviors. To do so, he and his team used behavioromics, recording and measuring individual aspects of mouse foraging behavior ranging from the size of an individual step to the specific destinations they stopped at, and then feeding the data through a machine learning algorithm.
Gregg and his team generated 12 groups of mice: males and females with either the maternal or paternal copy of both Th and Ddc knocked out, as well as heterozygous littermate controls for each gene. They then video recorded the mice as the animals made what Gregg calls “home-to-home foraging excursions,” in which they leave their enclosure and enter a connected arena designed to allow for natural behaviors such as exploring and digging in the sand for food before returning home.
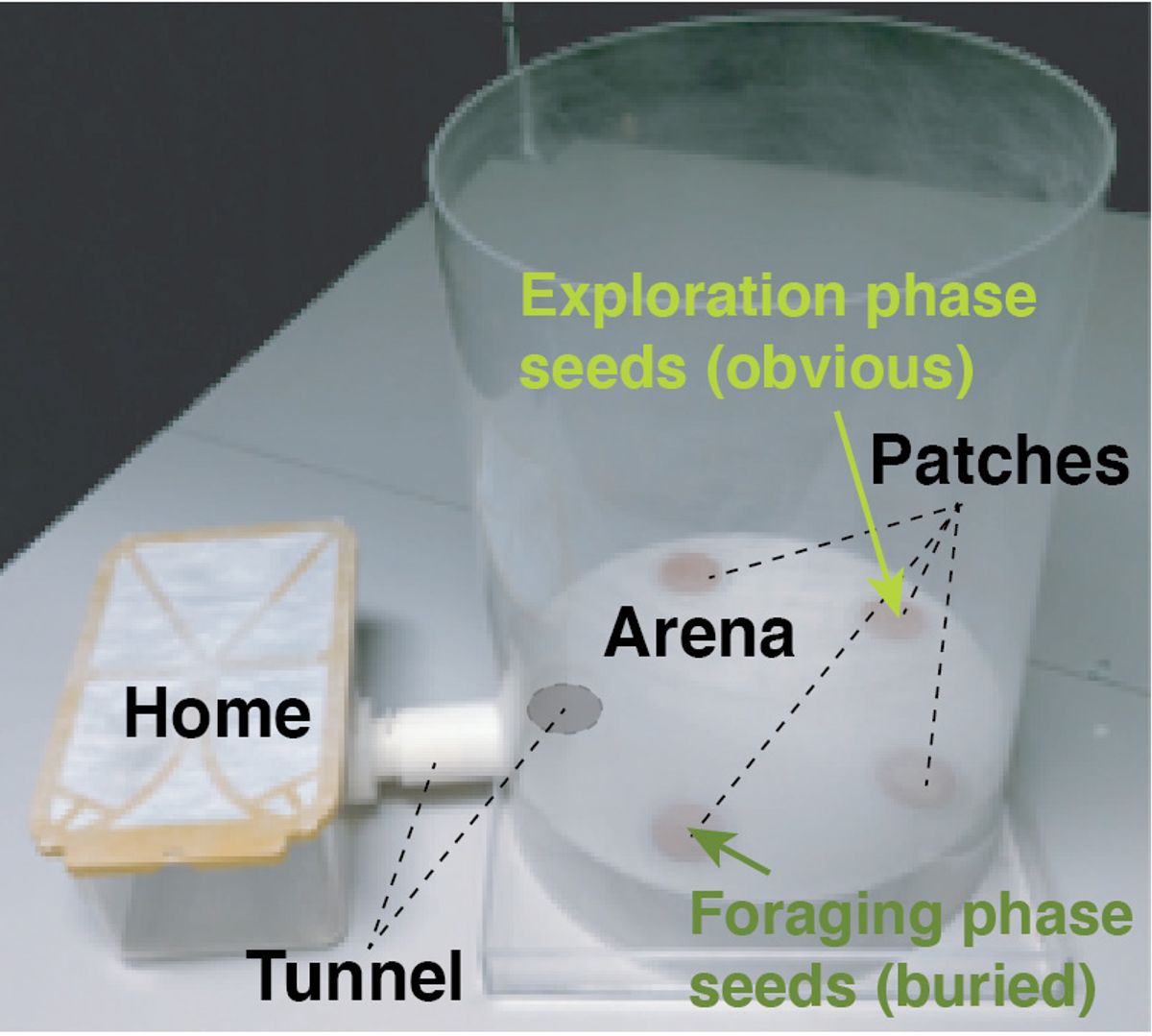
After 22,679 excursions by 426 mice from the experimental and control groups were complete, the team’s DeepFeats algorithm quantified a multitude of behavioral measures (“as much as we feasibly could,” says Gregg), and determined 16 measures that differed depending on which alleles were expressed.
Steven Millership, a genomics researcher at Imperial College London who didn’t work on the study, writes in an email to The Scientist that the paper impressed him. “[T]he authors appear to have taken considerable time to establish a foraging model whereby it is as close to the ‘wild’ as possible—although this is obviously very difficult to recreate in the lab research environment (and indeed they accept this),” he writes. “However, this kind of machine learning of phenotypic behavior can certainly be used in a kind of high throughput manner to tease out behavior that would otherwise be extremely difficult to detect by eye.”
The results indicate that male mice’s foraging behavior was altered when they lost the maternal allele of either gene. They also behaved differently if they lost the paternal allele of either gene, though fewer behaviors were affected and to a lesser extent. For example, males with their maternal Th or Ddc alleles knocked out traveled greater distances during their excursions, while males without their paternal alleles traveled the same distance as controls. Female mice only behaved differently to a significant extent when they lost their paternal alleles.
“The father’s copy of Ddc most strongly affected the decisions and actions that daughters make, and the mother’s copy most strongly affected the sons,” Gregg says. “In either case, the mom and dad’s alleles were affecting different actions and decisions.” Feil says he was surprised and fascinated by the finding of different effects in male and female mice, describing it as “pretty new.”
The results indicate that “there is some conflict between what the mothers and fathers would want their offspring to do or how they would behave,” Dulac says.
Feil says he hopes future studies will clarify the extent to which the effect differs among multiple mice with the same genes expressed—a distinction that will also reveal whether the imprinting arises randomly or if there’s some particular mechanism that, for example, “determines in this particular group of dopamine neurons that you get the maternal chromosome” that’s conserved from mouse to mouse.
“Whatever the mechanism is, it has to be pretty complex to explain these patterns,” Feil says.
It’s a beautiful demonstration that in some circumstances, at least, this parental bias corresponds to full silencing of some alleles.
—Catherine Dulac, Harvard University
Millership and Noordermeer both note that the study involved total knock out of the relevant alleles, meaning there may have been secondary effects caused by their absence in cells where both alleles would normally be expressed. Both say it would be interesting to see research that conditionally knocked out the genes only in the relevant cells.
The study authors also explored whether the preferential allele expression influenced physiology in addition to behavior, in this case focusing specifically on the activity of Ddc as expressed in each group of mutant mice. Gregg explains that for this gene, “If you lose the dad’s copy, then it affected dopamine levels in the urine of the daughters, but [there was] no effect if you lose the mom’s copy,” which was “consistent with dad influencing physiology and behavior in the daughters.” Conversely, in the male offspring, knocking out the maternal copy of Ddc affected dopamine levels, norepinephrine, and noradrenaline levels in urine, but no such effect was seen with the paternal allele.
How common is sexual dimorphism in imprinting?
Dulac and Gregg both say that an obvious next step is to repeat the process looking at any of the several other genes that show signs of imprinting. “How broad is this phenomenon?” Dulac wonders.
She also hopes to see researchers tackle the question of what impact this preferential, sexually dimorphic allele expression has at the population level, saying that it would be “really interesting” to find out what evolutionary benefits the phenomenon may confer.
“It’s a bit odd but there might be evolutionary reasons for which a paternal genome wants to influence the behavior of female offspring,” suggests Feil. He speculates that the sexually dimorphic imprinting effect and its influence on offspring behavior must have something to do with evolutionary success—offspring continuing to pass on the genome to the next generation—and that perhaps male mice developed a different evolutionary strategy for doing so than females.








